Building a Brazillian Jiu-Jitsu family tree
29 Dec 2016Martial arts masters are responsible for transmitting the skills and ideology of the art to their students. This master-student relationship is codified when a student receives his/her black belt, thereby becoming a master. The student’s future as a master of the art is forever tied to the person by whom he/she is promoted. Information in a martial art is transmitted through a sequence of master-student relationships. A master promotes students to masters, in turn, eventually promote students of their own.
While we normally think about the transference of information from masters to students, it is also informative to think about the flow of information as we move backwards in time. From this perspective, a student learns from his/her master, who learned from his/her master, and so on. This sequence of master-student relationships tracing back to the genesis of the art defines a martial artist’s lineage.
Here, I will visualize the lineages of hundreds of elite BJJ practitioners drawn from the website BJJ heroes.
I feel that lineages are intrinsically interesting; they allow BJJ practitioners, such as myself, to better understand our place in the context of an increasingly popular sport. Moreover, to the extent that style is transmitted through master-student relationships, practitioners who are nearby in a lineage-network are likely to be more stylistically similar than those from distant lineages.
Obtaining raw lineage data for ~800 BJJ heroes
To investigate BJJ fighter lineages, I used data from BJJ Heroes, a website containing biographies, lineages, fight records and more for around 800 elite BJJ practitioners. While these practitioners are among the more elite members of the sport (as defined by the BJJ Heroes crew), this list seems like a reasonable cross-section of the sport that is not greatly biased towards a subset of lineages or countries.
To extract lineages from this website, I first needed a list of all of the BJJ heroes that we are interested in. These fighters are conveniently located on a single page, so I could associate fighters’ names with links to their pages.
suppressPackageStartupMessages(library(dplyr))
suppressPackageStartupMessages(library(rvest))
library(tidyr)
strip_white <- function(x){
x %>%
gsub(pattern = '^[ ]+', replacement = '') %>%
gsub(pattern = '[ ]+$', replacement = '')
}
overwrite_cache <- FALSE
if (file.exists("BJJ_heroes_raw.Rdata") & overwrite_cache == FALSE) {
load("BJJ_heroes_raw.Rdata")
} else {
# ID all BJJ heroes
all_bjj_fighters <- xml2::read_html("http://www.bjjheroes.com/a-z-bjj-fighters-list")
# Save fighter summaries
all_bjj_fields <- all_bjj_fighters %>%
# extract fields of interest
rvest::html_nodes(".column-4 , .column-3, .column-2, .column-1") %>%
rvest::html_text() %>%
# format as matrix
matrix(ncol = 4, byrow = T)
colnames(all_bjj_fields) <- all_bjj_fields[1,]; all_bjj_fields <- all_bjj_fields[-1,]
all_bjj_fields <- all_bjj_fields %>%
as.data.frame(stringsAsFactors = F) %>%
dplyr::tbl_df() %>%
# strip leading and trailing white space
dplyr::mutate_each(dplyr::funs(strip_white)) %>%
dplyr::mutate(Full_name = paste(`First Name`, `Last Name`))
# Save fighter links
all_bjj_fields$Fighter_link <- all_bjj_fighters %>%
# extract links to fighter pages
rvest::html_nodes(".column-1 a") %>%
rvest::html_attr("href")
# If fighter links are duplicated only keep one
all_bjj_fields <- all_bjj_fields %>%
dplyr::group_by(Fighter_link) %>%
dplyr::slice(1) %>%
dplyr::ungroup()
# Generate absolute links (some links are relative, others are absolute)
all_bjj_fields <- all_bjj_fields %>%
dplyr::mutate(Fighter_link = ifelse(grepl("^http", Fighter_link), Fighter_link, paste0("http://www.bjjheroes.com", Fighter_link)))
}
knitr::kable(all_bjj_fields %>% dplyr::sample_n(5), row.names = F)| First Name | Last Name | Nickname | Team | Full_name | Fighter_link |
|---|---|---|---|---|---|
| Alan | Moraes | Carlson Gracie | Alan Moraes | http://www.bjjheroes.com/?p=2583 | |
| Adilson | Lima | Bitta | Academia Pitbull | Adilson Lima | http://www.bjjheroes.com/?p=6768 |
| Ricardo | Rezende | Fight Sports | Ricardo Rezende | http://www.bjjheroes.com/?p=4859 | |
| Marcos | de Souza | Bonsai | Marcos de Souza | http://www.bjjheroes.com/?p=1848 | |
| Maximiliano | Trombini | Cia Paulista | Maximiliano Trombini | http://www.bjjheroes.com/?p=872 |
After identifying all of the pages of BJJ heroes, I programmatically followed each hero’s link and saved the lineage field from each hero.
if (!file.exists("BJJ_heroes_raw.Rdata") | overwrite_cache == TRUE) {
# Crawl fighter pages
fighter_output <- list()
for(i in 1:nrow(all_bjj_fields)){
fighter_page <- try(read_html(all_bjj_fields$Fighter_link[i]), silent = T)
if(class(fighter_page)[1] == "try-error"){next} # catch missing pages
fighter_attr <- fighter_page %>% html_nodes("p:nth-child(4), p:nth-child(5), p:nth-child(6), p:nth-child(7), p:nth-child(8)") %>% html_text()
# match lineage field
fighter_lineage <- fighter_attr[grep('[lL]ineage', fighter_attr)][1] # only take on lineage
if(length(fighter_lineage) == 0){print(i)}else{
fighter_output[[i]] <- fighter_lineage
}
}
BJJ_lineages <- lapply(1:nrow(all_bjj_fields), function(i){
if(length(fighter_output[[i]]) != 0){
data.frame(all_bjj_fields %>%
slice(i),
Lineage = fighter_output[[i]],
stringsAsFactors = FALSE)
} # catch missing pages that were skipped with try-error
}) %>%
bind_rows()
save(all_bjj_fields, BJJ_lineages, file = "BJJ_heroes_raw.Rdata")
}
knitr::kable(BJJ_lineages %>% dplyr::sample_n(5), row.names = F)| First.Name | Last.Name | Nickname | Team | Full_name | Fighter_link | Lineage |
|---|---|---|---|---|---|---|
| Ana Carolina | Vieira | GF Team | Ana Carolina Vieira | http://www.bjjheroes.com/?p=6979 | Lineage: Mitsuyo Maeda > Luis França > Oswaldo Fadda > Monir Salomão > Julio Cesar > Ana Carolina Vieira | |
| Kayron | Gracie | Gracie Barra | Kayron Gracie | http://www.bjjheroes.com/?p=901 | Lineage: Mitsuyo Maeda > Carlos Gracie Sr. > Helio Gracie > Carlos Gracie Junior > Kayron Gracie | |
| Marcus | Bello | GF Team | Marcus Bello | http://www.bjjheroes.com/?p=1619 | Lineage: Mitsuyo Maeda > Luis França > Oswaldo Fadda> Monir Salomão > Julio Cesar Pereira > Marcus Bello | |
| Guilherme | Augusto | Alliance | Guilherme Augusto | http://www.bjjheroes.com/?p=5417 | Lineage: Mitsuyo Maeda > Carlos Gracie > George Gracie > Octávio de Almeida > Moises Murad > Everdan Olegário > Guilherme Augusto | |
| Gary | Tonon | Renzo Gracie Academy | Gary Tonon | http://www.bjjheroes.com/?p=5649 | Lineage: Mitsuyo Maeda > Carlos Gracie Sr. > Helio Gracie > Carlos Gracie Junior > Renzo Gracie > Ricardo Almeida (> Tom deBlass) > Garry Tonon |
Each hero’s lineage is stored as a string, like “lineage: master’s master > master > student” (with some other variations), that contains the overall lineage of each hero. It is useful to think about these lineages as networks where direct connections between masters and their students are parent-child relationships. Links that span multiple parent-child links are termed ancestor-descendent relationships.
Combining the lineages of all individual fighters into an overall family tree can be accomplished once I have identified all of the master-student relationships in this database.
Summarizing lineages based on master-student relationships
To identify the master-student relationships that will form individual branches of the BJJ family tree, I first need to unpack each hero’s lineage by removing extraneous text and storing each ancestor as a distinct field.
# Clean up and filter missing lineages
clean_lineage <- function(x){
# extract fields following "lineage"
x <- strsplit(x, split = '[Ll]ineage[A-Za-z0-9 )(]{0,}:[[:space:]]{0,}|(Main Achi)')[[1]][2]
# remove leading and trailing white space
gsub('[[:space:][:punct:]]+$', '', gsub('^[[:space:][:punct:]]+', '', x))
}
BJJ_lineages <- BJJ_lineages %>%
select(Fighter_link, Lineage) %>%
rowwise() %>%
mutate(Lineage = clean_lineage(Lineage))
# filter missing lineages
BJJ_lineage <- BJJ_lineages %>%
ungroup() %>%
filter(!is.na(Lineage)) %>%
filter(!(Lineage %in% c("", "—", "n/a")))
# Turn lineages into a hierarchy
BJJ_lineage_unpacked <- lapply(1:nrow(BJJ_lineage), function(i){
x <- strsplit(BJJ_lineage$Lineage[i], split = ">")[[1]]
x <- gsub('[[:space:][:punct:]]+$', '', gsub('^[[:space:][:punct:]]+', '', x))
data.frame(Fighter_link = BJJ_lineage$Fighter_link[i], Lineage = x, stringsAsFactors = FALSE)
}) %>%
bind_rows()
# Label hierarchy for each student
BJJ_lineage_unpacked <- BJJ_lineage_unpacked %>%
group_by(Fighter_link) %>%
mutate(Level = 1:n()) %>%
mutate(Lineage = gsub('Master ', '', Lineage))
knitr::kable(BJJ_lineage_unpacked %>% dplyr::ungroup() %>% dplyr::slice(1:10), row.names = F)| Fighter_link | Lineage | Level |
|---|---|---|
| http://www.bjjheroes.com/?p=1000 | Mitsuyo Maeda | 1 |
| http://www.bjjheroes.com/?p=1000 | Carlos Gracie | 2 |
| http://www.bjjheroes.com/?p=1000 | Carlson Gracie | 3 |
| http://www.bjjheroes.com/?p=1000 | Ze Mario Sperry | 4 |
| http://www.bjjheroes.com/?p=1000 | Caroline de Lazzer | 5 |
| http://www.bjjheroes.com/?p=1003 | Mitsuyo Maeda | 1 |
| http://www.bjjheroes.com/?p=1003 | Carlos Gracie | 2 |
| http://www.bjjheroes.com/?p=1003 | Helio Gracie | 3 |
| http://www.bjjheroes.com/?p=1003 | Francisco Mansor | 4 |
| http://www.bjjheroes.com/?p=1003 | Augusto Mendes | 5 |
Having separated each lineage into separate entry for each ancestor, these lineages can now be used to form master-student relationships. Master-student relationships may be shared by multiple descendents. For example: >400 lineages contain the master-student relationship between Mitsuyo Maeda and his student Carlos Gracie. Each of these shared master-student relationships only needs to be stored once (although I also keep track of how many descendents each fighter has).
# Identify all master-student relationships
master_student_relationships <- BJJ_lineage_unpacked %>%
rename(Master = Lineage) %>%
left_join(BJJ_lineage_unpacked %>%
rename(Student = Lineage) %>%
mutate(Level = Level - 1),
by = c("Fighter_link", "Level")) %>%
filter(!is.na(Student)) %>%
count(Master, Student, Level) %>%
ungroup() %>%
arrange(Level, desc(n))
# reduce lineage to one master per student (preferring the master with the most students)
master_student_relationships = master_student_relationships %>%
group_by(Student) %>%
arrange(desc(n)) %>%
mutate(n = sum(n)) %>%
slice(1) %>%
ungroup()
# At each level, combine very similar names of students
# root masters are handled seperately
root_masters <- master_student_relationships %>%
filter(Level == 1) %>%
select(Master, Level, n) %>%
count(Master, Level, wt = n) %>%
dplyr::rename(n = nn) %>%
ungroup() %>%
arrange(desc(n))
knitr::kable(master_student_relationships %>% dplyr::sample_n(10), row.names = F)| Fighter_link | Master | Student | Level | n |
|---|---|---|---|---|
| http://www.bjjheroes.com/?p=781 | Ricardo De La Riva | Rodrigo Nogueira | 4 | 1 |
| http://www.bjjheroes.com/?p=1577 | Leonardo Vieira | Rafael Heck | 6 | 1 |
| http://www.bjjheroes.com/?p=1604 | Leonardo Vieira | Nivaldo Oliveira | 6 | 1 |
| http://www.bjjheroes.com/?p=6046 | John Lewis | Gazzy Parman | 5 | 1 |
| http://www.bjjheroes.com/?p=6064 | John Lewis | Steve da Silva | 5 | 1 |
| http://www.bjjheroes.com/?p=4430 | Cesar Guimaraes | Fabiano Gaudio | 6 | 2 |
| http://www.bjjheroes.com/?p=2560 | Leoni Nascimento | Luiz Dias | 4 | 1 |
| http://www.bjjheroes.com/?p=2206 | Julio Lima | Sandro Lima | 5 | 1 |
| http://www.bjjheroes.com/?p=1174 | Kazuo Yoshida | Evaldo Luiz “Serrinha | 1 | 1 |
| http://www.bjjheroes.com/?p=7077 | Wilson Mattos | Manoel Costa | 4 | 1 |
Having aggregated all master-student relationships, I could move to visualizing the overlapping lineages of heroes; however, there are currently inconsistencies in the data that would muddy the results. This problem can be seen by looking at Mitsuyo Maeda, one of the founders of the sport who is an ancestor of almost all fighters in this dataset.
| Master | Level | n |
|---|---|---|
| Mitsuyo Maeda | 1 | 720 |
| M. Maeda | 1 | 13 |
| Takeo Iano | 1 | 6 |
| Kazuo Yoshida | 1 | 1 |
| Takeo Yano | 1 | 1 |
Most lineages refer to Mitsuyo Maeda as “Mitsuyo Maeda” but some list him as “M. Maeda.” We see a similar problem with another of the sport’s founders: “Takeo Yano,” also referred to as “Takeo Iano.” Without consolidating such records, some fighters with the same lineage would be artificially separated. In the case of Mitsuyo Maeda, we would need to allow for the insertion or removal of characters to achieve a match. For Takeo Yano, we would need to allow for the substitution of characters.
Dealing with inconsistent hero names is a tricky challenge; we want to match similar strings without inappropriately lumping heroes together. This is especially challenging given the extremely similar names within the Gracie family! With simple string matching approaches, it would be very difficult not to combine names like Rolls, Royce, Royler and Renzo Gracie, while also still appropriately combining alternative spellings such as “Luis Franca” with “Luiz França”.
To combine a set of fighter names, I relied upon a few rules:
- Names can only be combined if they have the same master. (These master names may have already been merged.)
- Names that exactly match the BJJ heroes’ entries that I found above are first matched. (This deals with the Gracies’ similar names.)
- Unmatched names are sorted according to how many ancestors they have.
- Each unmatched name is tested to see if it matches one of the earlier student names (rule 3) using either of two approaches:
- Maximum subsequence: to identify insertions or deletions, I see how many characters the unmatched name has in common with each previous student and normalize this subsequence to the length of the shortest string. This approach is good at catching abbreviations such as M. Maeda.
- Levenshtein distance: to identify alternative spellings, I determine how many characters need to be either inserted, deleted or changed to another character in order to turn one name into another. This approach is good at combining alternative spellings of a name such as with Luis Franca.
- Renamed athletes are manually checked and renaming is overruled when appropriate, using a list of manual corrections.
In addition to alternative names, some individuals with similar lineages list different master-student relationships in their lineages. To make sense of these inconsistencies, I assumed that each student has a single master (choosing the master with the most descendents). This simplification neglects that some students should legitimately have multiple masters, but by enforcing a one-to-many master-to-students relationship, I impose a strict hierarchy on the lineages that will improve clarity later on.
Before broadly combining names, I first wrote functions for aggregating names. I applied these functions to the root masters to resolve the inconsistent naming of Mitsuyo Maeda and Takeo Yano.
# Combine alternative names
LCS_wrap <- function(x,y){
# maximum subsequence between one x and one y (normalized relative to the shorter string)
# low values are best
LCS_obj = qualV::LCS(strsplit(tolower(x), split = '')[[1]], strsplit(tolower(y), split = '')[[1]])
1 - LCS_obj$LLCS / min(length(LCS_obj$a), length(LCS_obj$b))
}
LEV_frac <- function(x,y){
# levenshtein distance normalized to max cost
match_costs <- list(insertions = 4, deletions = 4, substitutions = 1)
x <- tolower(x); y <- tolower(y)
str_lengths <- sapply(y, function(z){strsplit(z, split = '')[[1]] %>% length()})
str_lengths <- sapply(str_lengths, max, length(strsplit(x, split = '')[[1]]))
adist(x, y, costs = match_costs) / (str_lengths*max(unlist(match_costs)))
}
combine_names <- function(x, counts, priority_names, overwrites){
# provide a vector names with the highest categories first
# returns names of later names that match early names
# Two possible ways of matching:
# long common subsequence (to catch abbreviations)
# few letter substitutions
name_overwrite = data.frame(old = x, counts = counts,
score_indel = NA, score_sub = NA, stringsAsFactors = FALSE) %>%
# overwrite old with new overwrite (if provided)
left_join(overwrites, by = "old") %>%
# prioritize names in priority_names (i.e. generally those in the BJJ heroes database verbatim)
mutate(new = ifelse(old %in% priority_names, old, new),
input_order = 1:n()) %>%
arrange(new, desc(counts))
if(all(is.na(name_overwrite$new))){
name_overwrite$new[1] <- name_overwrite$old[1]
}
if(nrow(name_overwrite) == 1 | all(!is.na(name_overwrite$new))){
return_list <- list()
return_list$names <- name_overwrite$new[name_overwrite$input_order]
return_list$overwrites <- data.frame(old = c(), new = c(), stringsAsFactors = FALSE)
return(return_list)
}
for(i in (sum(!is.na(name_overwrite$new))+1):length(x)){
similarity_scores <- data.frame(old = name_overwrite$old[1:(i-1)], indel_diff = sapply(name_overwrite$old[1:(i-1)], LCS_wrap, y = name_overwrite$old[i]), sub_diff = c(LEV_frac(name_overwrite$old[i], name_overwrite$old[1:(i-1)])), stringsAsFactors = FALSE)
similarity_scores <- similarity_scores %>%
filter(indel_diff < 0.2 | sub_diff < 0.2)
if(nrow(similarity_scores) == 0){
name_overwrite$new[i] <- name_overwrite$old[i]
}else{
similarity_scores <- similarity_scores %>%
rowwise() %>%
mutate(best_score = min(indel_diff, sub_diff)) %>%
arrange(best_score) %>%
slice(1)
name_overwrite$new[i] <- name_overwrite$new[name_overwrite$old == similarity_scores$old]
name_overwrite$score_indel[i] <- similarity_scores$indel_diff
name_overwrite$score_sub[i] <- similarity_scores$sub_diff
}
}
return_list <- list()
return_list$names <- name_overwrite$new[order(name_overwrite$input_order)]
return_list$overwrites <- name_overwrite %>%
filter(old != new)
return_list
}
manual_renames <- tibble::frame_data(~old, ~new,
"Carlinhos Gracie", "Carlos Gracie Junior",
"R. Gracie", "Rickson Gracie",
"Ricardo Libório", "Ricardo Liborio",
"Cássio Cardoso", "Cassio Cardoso",
"Luis Duarte", "Luis Roberto Duarte",
"Marcus Silveira", "Marcus Silveira",
"Vinicius Magalhães", "Vinicius Magalhaes",
"Toni Pontes", "Toni Pontes",
"Laerte Barcelos", "Laerte Barcelos",
"Sergio Lisboa", "Sergio Lisboa",
"Helio Soneca", "Helio Soneca",
"Roberto Maia", "Roberto Maia",
"Daniel Simoes “Gracie”", "Daniel Simoes",
"José Leão Teixeira", "Jose Leao Teixeira",
"Rafael Fofitio", "Rafael Barros",
"Renan Barao", "Renan Pegado",
"Binho Monteiro", "Fabio Monteiro",
"Richie Martinez", "Richard Martinez",
"Geo Martinez", "Geovanny Martinez",
"L. Dalla", "Leonardo Dalla",
"Luis Gustavo", "Luis Gustavo",
"L. Irvin", "Lloyd Irvin",
"João Miyao", "Joao Miyao",
"Tiago Barros", "Hiago George",
"Jacare", "Romero Cavalcanti")
# similar names of the root masters (heroes who do not have a master listed)
agg_names <- combine_names(root_masters$Master, counts = root_masters$n, priority_names = all_bjj_fields$Full_name, overwrites = manual_renames)
knitr::kable(agg_names[["overwrites"]], row.names = F)| old | counts | score_indel | score_sub | new | input_order |
|---|---|---|---|---|---|
| M. Maeda | 13 | 0.125 | 0.4038462 | Mitsuyo Maeda | 2 |
| Takeo Yano | 1 | 0.100 | 0.0250000 | Takeo Iano | 5 |
Using this approach, the variants of Mitsuyo Maeda and Takeo Yano were combined. Carrying out such string matching across all fighters simultaneously would inappropriately group together a large number of names. Instead, I separately combined the students of each master using the criteria outlined above, sequentially moving to lower levels of the lineage hierarchy once higher level master names were aggregated. I also kept track of all of the combined names for manual verification and correction (to ensure that I did not do anything too heretical).
track_name_overwrites <- list()
track_name_overwrites[[1]] <- data.frame(Master = NA, agg_names[["overwrites"]], stringsAsFactors = FALSE) # track old and new names to vet namechanges
root_masters <- root_masters %>%
mutate(name_overwrite = agg_names[["names"]])
# Add back new names and reweight to aggregate combined names
master_student_relationships <- master_student_relationships %>%
left_join(root_masters %>%
select(-n),
by = c("Master", "Level")) %>%
mutate(Master = ifelse(!is.na(name_overwrite), name_overwrite, Master)) %>%
count(Master, Student, Level, wt = n) %>%
dplyr::rename(n = nn) %>%
ungroup() %>%
arrange(Level, desc(n))
# Carryout a similar process for each level of the hierarchy
# look at the students of each master and consolidate very similar names
for(a_level in sort(unique(master_student_relationships$Level))){
# query the master-student relationships for a single level of the hierarchy
master_students_level <- master_student_relationships %>%
filter(Level == a_level)
name_updates <- list()
# for each master, combine really similar student names
for(a_master in unique(master_students_level$Master)){
agg_names <- combine_names(master_students_level$Student[master_students_level$Master == a_master],
counts = master_students_level$n[master_students_level$Master == a_master],
priority_names = all_bjj_fields$Full_name,
overwrites = manual_renames)
if(nrow(agg_names[["overwrites"]]) != 0){
track_name_overwrites[[length(track_name_overwrites)+1]] <- data.frame(Master = a_master, agg_names[["overwrites"]], stringsAsFactors = FALSE)
}
name_updates[[length(name_updates)+1]] <- master_students_level %>%
filter(Master == a_master) %>%
mutate(name_overwrite = agg_names[["names"]])
}
updated_student_names <- name_updates %>%
bind_rows()
# Update master student relationships to reflect name changes and reweight to account for combined names
master_student_relationships <-
# overwrite student name slot
master_student_relationships %>%
left_join(updated_student_names %>%
select(-n),
by = c("Master", "Student", "Level")) %>%
mutate(Student = ifelse(!is.na(name_overwrite), name_overwrite, Student)) %>%
select(-name_overwrite) %>%
# overwrite masters name slot: the student now becomes the master (for the next iteration/level)
left_join(updated_student_names %>%
select(-Master, -n) %>%
rename(Master = Student) %>%
mutate(Level = Level + 1), by = c("Master", "Level")) %>%
mutate(Master = ifelse(!is.na(name_overwrite), name_overwrite, Master)) %>%
# recount with updated labels
count(Master, Student, Level, wt = n) %>%
dplyr::rename(n = nn) %>%
ungroup() %>%
arrange(Level, desc(n))
}
# overwrite additional names that were removed during the fuzzy matching (this process can be off if there are extra steps in some lineages with the same members
name_overwrites <- track_name_overwrites %>%
bind_rows()
knitr::kable(head(name_overwrites, 10), row.names = F)| Master | old | counts | score_indel | score_sub | new | input_order |
|---|---|---|---|---|---|---|
| NA | M. Maeda | 13 | 0.1250000 | 0.4038462 | Mitsuyo Maeda | 2 |
| NA | Takeo Yano | 1 | 0.1000000 | 0.0250000 | Takeo Iano | 5 |
| Mitsuyo Maeda | Carlos Gracie Sr | 133 | 0.0000000 | 0.1875000 | Carlos Gracie | 2 |
| Mitsuyo Maeda | Luiz França | 17 | 0.0909091 | 0.0227273 | Luis França | 4 |
| Mitsuyo Maeda | C. Gracie | 13 | 0.1111111 | 0.3269231 | Carlos Gracie | 5 |
| Mitsuyo Maeda | Carlos Gracie sr | 13 | 0.0000000 | 0.1875000 | Carlos Gracie | 6 |
| Mitsuyo Maeda | Luis Franca | 5 | 0.0909091 | 0.0227273 | Luis França | 7 |
| Mitsuyo Maeda | C. Gracie Sr | 1 | 0.0000000 | 0.2500000 | Carlos Gracie | 8 |
| Mitsuyo Maeda | Carlos Gracie/Helio Gracie | 1 | 0.0000000 | 0.5000000 | Carlos Gracie | 9 |
| Carlos Gracie | Carlinhos Gracie | 1 | NA | NA | Carlos Gracie Junior | 17 |
Since forming the original master-student relationships, I combined the names of some fighters and assigned each student to a single master. Because of the original lineage ambiguities, some masters may currently reside at multiple levels of our lineages. For example, Rickson Gracie is listed as a student of Carlos Gracie in some lineages and of Helio Gracie, in others. Because Helio is a student of Carlos, Rickson occurs on multiple lineage levels. In order to correct this problem, I releveled the lineage based on consensus master-student relationships so the lineages will form a proper hierarchy.
Once this is done, I have a set of unambiguous 849 master-student relationships that will construct the BJJ family tree.
ronin <- master_student_relationships %>%
filter(Master %in% setdiff(master_student_relationships$Master, master_student_relationships$Student))
ronin <- ronin %>%
left_join(name_overwrites %>%
select(Master = old, new),
by = "Master") %>%
filter(!is.na(new)) %>%
select(Master, Student, new)
master_student_relationships <- master_student_relationships %>%
left_join(ronin, by = c("Master", "Student")) %>%
mutate(Master = ifelse(!is.na(new), new, Master)) %>%
select(-new) %>%
# if a fighter has 2 masters (only possible if they showed up on different levels of the hierarchy, take the master who is more commonly accepted
dplyr::group_by(Student) %>%
dplyr::arrange(desc(n)) %>%
dplyr::slice(1) %>%
dplyr::ungroup()
# Relevel to ensure an unambiguous sequence from root masters to leaves
releveled_list <- list()
i <- 1
repeat{
if(i == 1){
releveled_list[[i]] <- master_student_relationships$Master[master_student_relationships$Level == 1] %>%
unique()
}else{
releveled_list[[i]] <- master_student_relationships %>%
dplyr::filter(Master %in% releveled_list[[i-1]]) %>%
dplyr::select(Student) %>%
unlist() %>%
unname()
if(length(releveled_list[[i]]) == 0){break}
}
i <- i+1
}
lineage_levels <- lapply(1:length(releveled_list), function(x){
if(length(releveled_list[[x]]) != 0){
data.frame(Student = releveled_list[[x]], Level = x-1, stringsAsFactors = FALSE)
}}) %>%
dplyr::bind_rows()
# relevel the lineages
master_student_relationships <- master_student_relationships %>%
dplyr::select(-Level) %>%
dplyr::left_join(lineage_levels, by = "Student")Static BJJ family tree
To first visualize the BJJ family tree, I was interested in generating a plot that showed all students in a hierarchical network, emphasizing masters who have a large number of descendents. Here, the founders of the sport are located in the middle of the plot, and students form sequential layers on the outside of their masters. When I originally generated this plot, I did not have a good tool for visualizing hierarchical data that falls in discrete levels (in retrospect, data.tree is probably the way to go), so I wrote a method for generating the layout from scratch.
a_lineage <- master_student_relationships
# Generate a manual dendrogram of lineages
# a linear order of individuals forms x
# y is found based on the maximum level of descendents
# form 1.1.2 type orderings for each fighter - order after each split
ordered_lineage <- a_lineage %>%
ungroup() %>%
arrange(Level, desc(n))
master_order <- ordered_lineage %>%
filter(Level == 1) %>%
count(Master, wt = n) %>%
dplyr::rename(n = nn) %>%
arrange(desc(n)) %>%
mutate(Order_str = 1:n())
ordered_lineage <- ordered_lineage %>%
left_join(master_order %>%
select(-n),
by = "Master")
for(i in sort(unique(ordered_lineage$Level))){
student_order <- ordered_lineage %>%
filter(Level == i) %>%
group_by(Master) %>%
arrange(desc(n)) %>%
mutate(updated_Order_str = paste(Order_str, 1:n(), sep = "."))
# Update students
ordered_lineage <- ordered_lineage %>%
left_join(student_order %>%
select(-n, -Order_str),
by = c("Level", "Master", "Student")) %>%
mutate(Order_str = ifelse(is.na(updated_Order_str), Order_str, updated_Order_str)) %>%
select(-updated_Order_str)
# Update the masters
ordered_lineage <- ordered_lineage %>%
left_join(student_order %>%
ungroup() %>%
select(Master = Student, Level, updated_Order_str) %>%
mutate(Level = Level + 1),
by = c("Master", "Level")) %>%
mutate(Order_str = ifelse(is.na(updated_Order_str), Order_str, updated_Order_str)) %>%
select(-updated_Order_str)
}
# Students (who are not masters) will form the bottom layer and their x coordinates will guide their master's layout
# Order students according to splits: e.g. 1.1.2
# Expand the individual nodes
exterior_students <- ordered_lineage %>%
filter(Student %in% setdiff(ordered_lineage$Student, ordered_lineage$Master))
student_tiers <- lapply(1:nrow(exterior_students), function(i){
entries <- strsplit(exterior_students$Order_str[i], split = "\\.")[[1]]
data.frame(Order = entries, Level = 1:length(entries), Order_str = exterior_students$Order_str[i], Student = exterior_students$Student[i], max.Level = exterior_students$Level[i], stringsAsFactors = FALSE)
}) %>%
bind_rows()
student_tiers <- student_tiers %>%
mutate(Order = as.numeric(Order)) %>%
spread(key = Level, value = Order) %>%
arrange_(.dots = paste0('`', as.character(sort(unique(student_tiers$Level))), '`')) %>%
mutate(x = 1:n())
all_masters <- rbind(
# root masters
data.frame(Master = ordered_lineage$Master[ordered_lineage$Level == 1],
Level = 0,
Order_str = substr(ordered_lineage$Order_str[ordered_lineage$Level == 1], 1, 1), stringsAsFactors = FALSE) %>%
group_by(Order_str) %>%
slice(1) %>%
ungroup(),
# masters who are students
ordered_lineage %>%
filter(Student %in% intersect(ordered_lineage$Student, ordered_lineage$Master)) %>%
select(Master = Student, Level, Order_str)
) %>%
arrange(desc(Level)) %>%
mutate(x = NA)
# masters are positioned by the mean of their root descendants
all_masters$x <- sapply(1:nrow(all_masters), function(i){
master_matches <- grep(paste0("^", all_masters$Order_str[i], "\\."), student_tiers$Order_str)
if(length(master_matches) > 1){
if(!all(diff(master_matches) == 1)){
stop(i)
}
}
mean(student_tiers$x[master_matches])
})
all_masters$n_students <- sapply(1:nrow(all_masters), function(i){
master_matches <- grep(paste0("^", all_masters$Order_str[i], "\\."), student_tiers$Order_str)
length(master_matches)
})
# Figure out y coordinates
find_lowest_level_descendent <- function(ordered_lineage, master){
starting_desc <- ordered_lineage %>% filter(Master == master)
if(nrow(starting_desc) == 0){return(0)}
current_students <- starting_desc$Student
repeat{
if(sum(ordered_lineage$Master %in% current_students) == 0){
break
} else {
current_students <- ordered_lineage$Student[ordered_lineage$Master %in% current_students]
}
}
ordered_lineage$Level[ordered_lineage$Student %in% current_students][1]
}
max_height = 10
all_masters <- all_masters %>%
arrange(Level) %>%
mutate(y = NA)
for(i in 1:nrow(all_masters)){
if(all_masters$Level[i] == 0){all_masters$y[i] <- 0; next}
masters_master <- ordered_lineage$Master[ordered_lineage$Student == all_masters$Master[i]]
# find the maximum number of tiers below the master (including his slot)
levels_below <- find_lowest_level_descendent(ordered_lineage, all_masters$Master[i]) - all_masters$Level[i] + 1
height_change <- (max_height - all_masters$y[all_masters$Master == masters_master]) / levels_below
all_masters$y[i] <- all_masters$y[all_masters$Master == masters_master] + height_change
}
# link descendents to ancestors
all_students <- student_tiers %>%
select(Fighter = Student, Level = max.Level, Order_str, x) %>%
mutate(y = max_height)
all_masters <- all_masters %>%
rename(Fighter = Master)
non_root_fighters <- rbind(all_students,
all_masters %>%
select(-n_students)) %>%
arrange(Level)
edge_list <- lapply(non_root_fighters$Fighter[non_root_fighters$Level != 0], function(x){
fighter_pos <- c(x = non_root_fighters$x[non_root_fighters$Fighter == x], y = non_root_fighters$y[non_root_fighters$Fighter == x])
master <- ordered_lineage$Master[ordered_lineage$Student == x]
master_pos <- c(x = non_root_fighters$x[non_root_fighters$Fighter == master], y = non_root_fighters$y[non_root_fighters$Fighter == master])
rbind(
c(xstart = fighter_pos['x'], xend = fighter_pos['x'], ystart = fighter_pos['y'], yend = master_pos['y']),
c(xtart = fighter_pos['x'], xend = master_pos['x'], ystart = master_pos['y'], master_pos['y'])
) %>% as.data.frame(stringsAsFactors = FALSE)
}) %>%
bind_rows()
suppressPackageStartupMessages(library(ggrepel))
ggplot() +
geom_segment(data = edge_list, aes(x = xstart.x, xend = xend.x, y = ystart.y, yend = yend.y), color = "#F8C300") +
geom_point(data = all_students, aes(x = x, y = y, name = Fighter), color = "#00923F", size = 1) +
geom_point(data = all_masters, aes(x = x, y = y, size = log2(n_students)), color = "#00923F", fill = "#28166F", shape = 21) +
geom_label_repel(data = all_masters %>% filter(n_students > 8), aes(x = x, y = y, label = Fighter),
label.padding = grid::unit(0.1, "lines"), color = "#28166F", size = 6) +
theme_void() + theme(legend.position = "none") +
coord_polar() +
scale_y_continuous(limits = c(-0.5, max_height)) +
scale_size_continuous(range = c(1,8))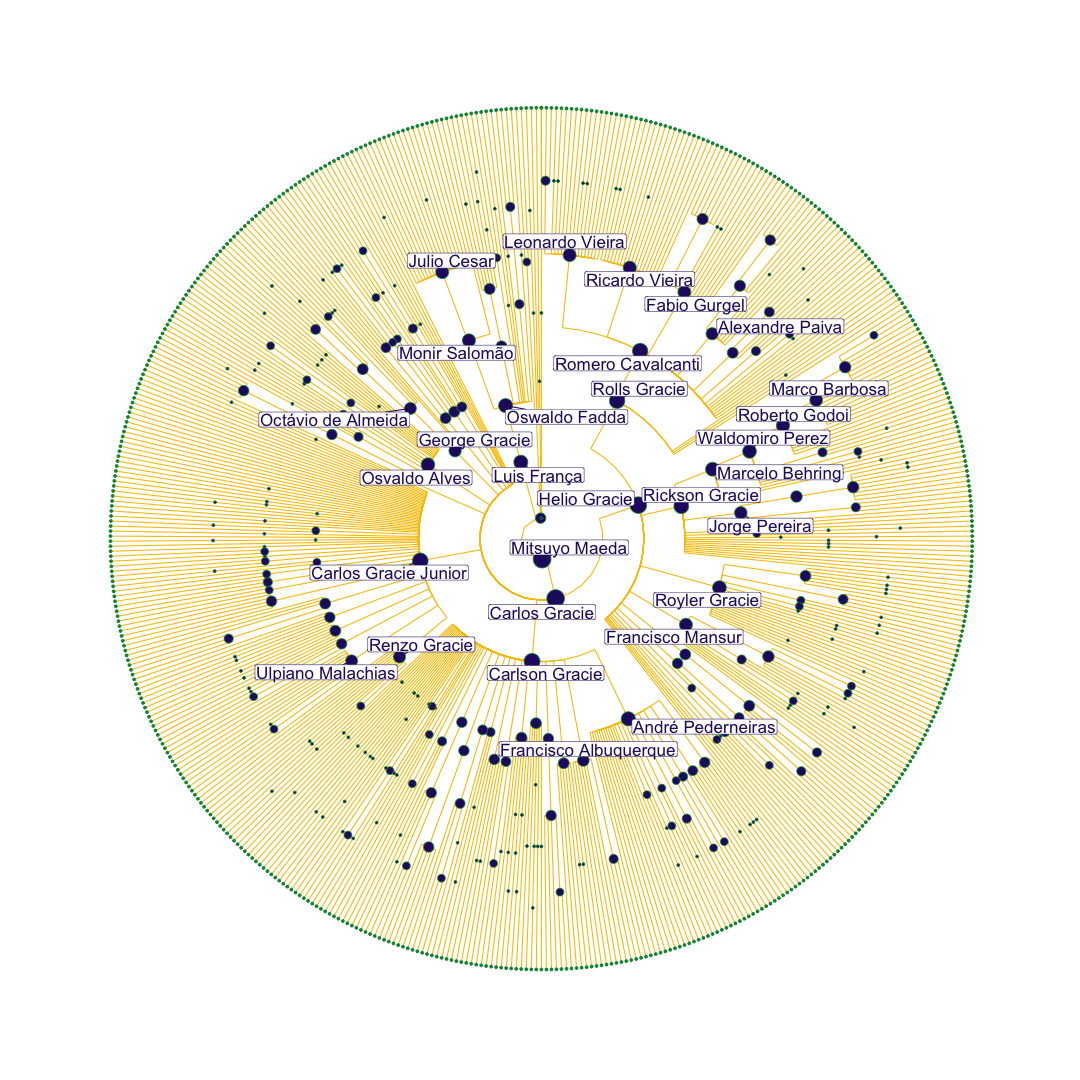
Interactive BJJ family tree
While the above static plot is useful for understanding the broad divisions in BJJ, I was only able to show about 30 heroes and still maintain legibility. In order to visualize the lineages of over 800 fighters, I needed an approach that allowed people to focus on subsets of the network. To achieve this goal, I used networkD3, which provides methods for generating interactive hierarchical radial plots using D3. I then added a CSS to alter the plot’s appearance. This interactive plot (each master’s students can be collapsed by clicking on the master) can be found here.
# reform each leaf student (student who is not a master) from consensus athletes
root_masters <- setdiff(master_student_relationships$Master, master_student_relationships$Student)
master_student_relationships <- dplyr::bind_rows(
#data.frame(Master = NA_character_, Student = "ALL", Level = 0, n = 0, stringsAsFactors = FALSE),
master_student_relationships %>%
filter(Level == 1) %>%
count(Master, Level, wt = n) %>%
dplyr::rename(n = nn) %>%
rename(Student = Master) %>%
mutate(Master = "ALL"),
master_student_relationships)
leafs <- setdiff(master_student_relationships$Student, master_student_relationships$Master)
leaf_paths <- lapply(leafs, function(leaf) {
path <- leaf
repeat {
match_master <- master_student_relationships$Master[master_student_relationships$Student == path[length(path)]]
if (length(match_master) > 1) {
stop("practicioner should not have multiple masters, structural problem in analysis")
} else if (length(match_master) == 1){
path <- c(path, match_master)
} else {
return(tibble::data_frame(leaf = leaf, practicioner = path) %>% dplyr::mutate(step = n():1))
}
}
}) %>%
dplyr::bind_rows()
BJJ_consensus_lineages <- leaf_paths %>%
dplyr::group_by(leaf) %>%
dplyr::arrange(leaf, step) %>%
dplyr::group_by(leaf) %>%
dplyr::summarize(pathString = paste(practicioner, collapse = "--"))
BJJ_tree <- BJJ_consensus_lineages %>%
data.tree::as.Node(pathDelimiter = "--") %>%
data.tree::ToListExplicit(unname = TRUE)
d3_bjj_tree <- networkD3::hierNetwork(BJJ_tree, type = "tree.radial",
zoomable = T, collapsible = T,
height = 2000, width = 2000, radius = 3, margin = 0)
networkD3::saveNetwork(d3_bjj_tree, file = "BJJfamilytree.html")